Background and Research Rationale:
Research description
Complex interactions between environmental factors, metabolic intermediates, commensal microbiota and genetic traits underpin the etiology of many infectious and immune-mediated diseases as well as the functioning of the immune system. Consequently, disrupted communication circuits between the host and the intestinal microbiota hamper the physiologic functions of various organ systems and insult the integrity of adjacent and distant tissues. In order to restore organ physiology and tissue integrity, our laboratory has been studying the mechanisms and pathways underlying the maintentance of mucosal immune tolerance and colonization resistance. Therefore, we have been investigating the pathogenesis and virulence of mucosal pathogens and have been developing vaccination strategies that inhibit the colonization with and the outgrowth of pathobiontic and pathogenic bacteria. Moreover, we have been analyzing the consequences of disupted host-microbiota interactions and genetic polymorphisms on immune-mediated diseases of the gut, the liver, the lung and the pancreas and have developed respective interventional strategies for prevention and cure.
Bacterial pathogenesis and vaccination
Bacterial virulence factors and functional as well as structural characteristics of the host and its immune response promote the attachment to and the invasion of mucosal tissue sites by various pathogens. Due to their well-documented role in the adhesion to the intestinal epithelium and their structural stability, we could identify bacterial surface glycans of Clostridioides difficile (C. diff.), the most frequent cause of antibiotic associated diarrhea, as suitable targets for vaccination. Currently, we are studying the mode of action of the protective antibody response in the inhibition of C. diff. colonization, translate our findings into clinical studies and attempt to identify suitable targets for vaccination against other gastrointestinal pathogens, such as Salmonella Typhimurium or Shigella flexneri. To understand the role of identified vaccination targets in bacterial pathogenesis and virulence, we are also studying deletional mutant strains of respective bacteria.
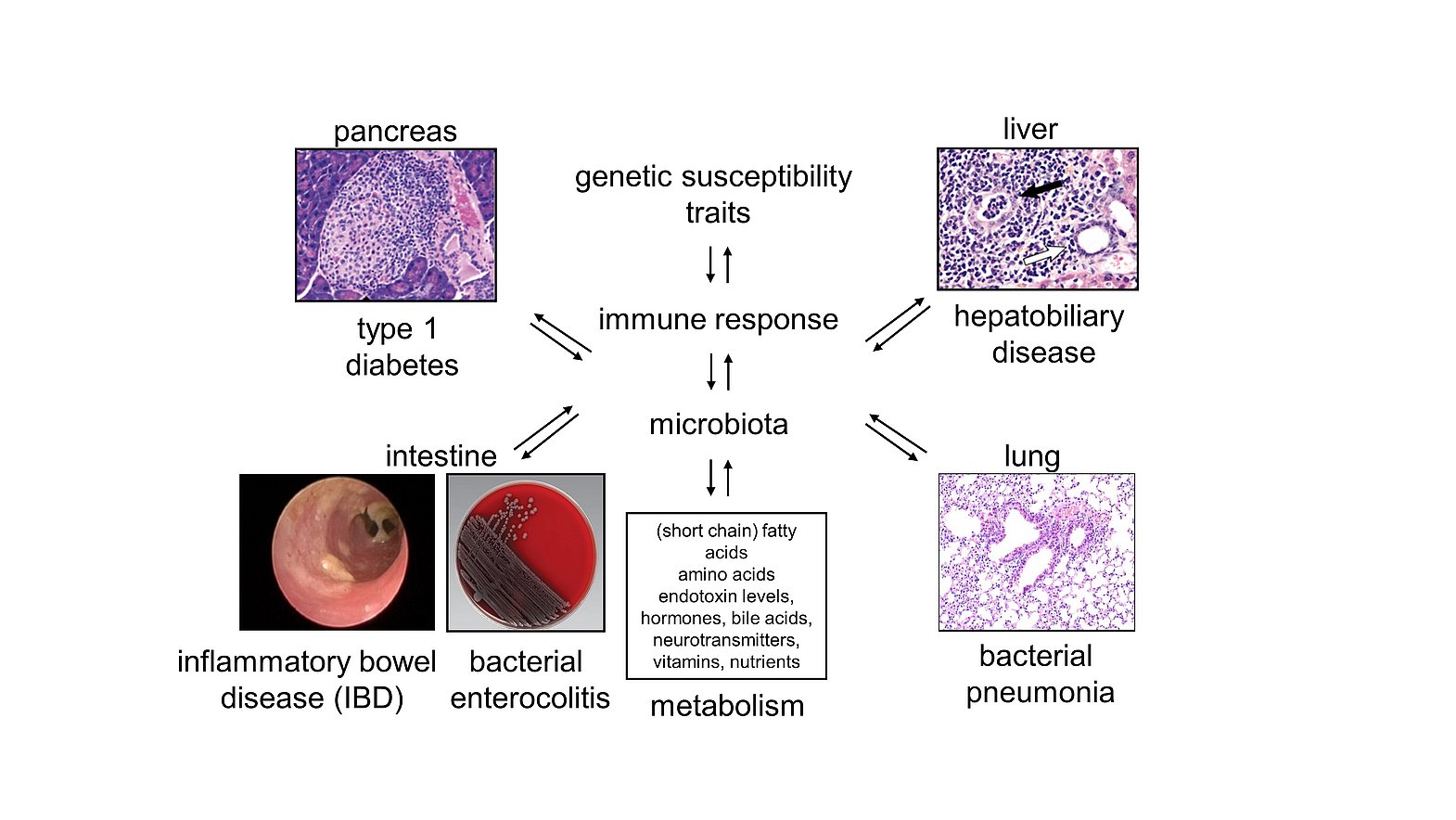
Host-microbiota cross-talk and inter-organ communication
Tissue-resident microbiota interfere with the host in multiple ways. Intestinal microbiota, for example, promote or inhibit inflammatory and regulatory pathways by signalling through different receptors or altering the intraluminal metabolome. Moreover, commensal microbiota prevent pathogenic bacteria from colonizing mucosal tissues such as the gastrointestinal tract or the lung, a phenomenon known as colonization resistance, due to the competition for nutrients and/or specific receptors and the regulation of the local immune response as well as the biophysical properties of the (muco-)epithelial barriers. Thereby inflammatory responses can affect local and distanct tissues as the gut micromilieu can prime immune cells that infiltrate extra-intestinal tissues. Intestinal microbiota also communicate with these tissue sites due to the release of extracellular vesicles and various organ systems can bidirectionally communicate with each other due to the mutual exchange of cytokines, chemokines, extracellular vesicles, metabolites and/or hormones. In this context, we are studying the interactions of intestinal bacteria, their metabolites and the vesicles that they can release in the maintenance of colonization resistance and immune tolerance in the gut, the liver, the lung and the pancreas. We have identified in this context the semi-essential amino acid L-arginine and its versatile metabolic pathways as promising targets for clinical intervention in colitis.
Genetic susceptibility to infections and immune-mediated disease
Inflammatory processes in response to microbial or self-antigens result from complex interactions of genetic, predisposing factors, antigens derived from the intestinal microbiota and distinct environmental cues. Some genetic regions are even associated with susceptibility to multiple immune-mediated disorders. In addition, inflammatory processes are frequently tissue-specific, although (auto-)antigens targeted by the immune system can be expressed throughout the entire body. Using different genetic approaches and immune- or infection- triggered models, we identified, for example, allelic polymorphisms within an immunoglobulin-like glycoprotein named CD101 as modifiers of immune-mediated and infection driven inflammatory disease. While genetic CD101 polymorphisms mediated protection from spontaneous type 1 diabetes, they enhanced susceptibility to infection-triggered colitis and liver pathology. Thus, CD101 is an example for an allele, which orchestrates a remarkable switch to tissue-specific pathology once specific bacteria are present in the respective micromilieus. Further studies are delineating the mechanisms that regulate the expression and function of CD101 and assess whether CD101 is a suitable biomarker for disease and/or clinical intervention.