Molecular Biology of Malaria Parasites
Malaria is caused by protozoan pathogens that are transmitted through the bite of an infected Anopheles mosquito. Malaria pathogenesis relies on several critical processes that represent Achilles heels of the parasite with great potential as targets for therapeutic interventions and vaccine development. Our group aims to identify novel targets for therapies against the human malaria parasite Plasmodium falciparum by investigating molecular mechanisms that contribute to the regulation of genes driving malaria pathogenesis and parasite differentiation. We apply state-of-the-art technologies including genome-wide RNA/DNA/Chromatin sequencing, quantitative proteomics approaches, high resolution imaging, and emerging genome editing tools to functionally and mechanistically investigate the molecular players that control chromatin based gene regulation in P. falciparum. Our research program focuses on the transcriptional regulation of three processes that are critical in the malaria life cycle:
1) Erythrocyte invasion. During its asexual life cycle in the human host, the malaria parasite relies on the invasion of and multiplication in erythrocytes. Interfering with the ability of the parasite to invade erythrocytes has been a focus of vaccine development over the past decades. We have recently identified a chromatin protein (PfBDP1) in Plasmodium falciparum that is essential for the parasite’s ability to transcribe invasion ligands at the correct point in time. Consequently, depletion of PfBDP1 results in a dramatic reduction of erythrocyte invasion (Josling et al., Cell Host & Microbe 2015). We aim to further investigate the molecular mechanisms by which PfBDP1 regulates the coordinate expression of invasion ligands.
2) Antigenic variation. To evade the host immune response, malaria parasites employ antigenic variation of parasite proteins that are exported to the host cell surface. While in recent years we have developed a good grasp of how the majority of genes coding for different variants are maintained in a repressed state through heterochromatin, it is still poorly understood how the activation of a single variant is achieved (Duffy et al., Brief. Funct. Genomics 2014). Our research has implicated novel histone variants in the active transcription of variant surface antigens (Petter et al., PLoS Pathogens 2011, Petter et al., Molecular Microbiology 2013). By combining transgenic approaches and genome wide sequencing with cellular and biochemical assays, we investigate the events leading to the deposition of histone variants and other activating marks at the transcriptional start site of the genes coding for variant surface antigens. This research aims to uncover novel aspects of the switching machinery directing antigenic variation in P. falciparum.
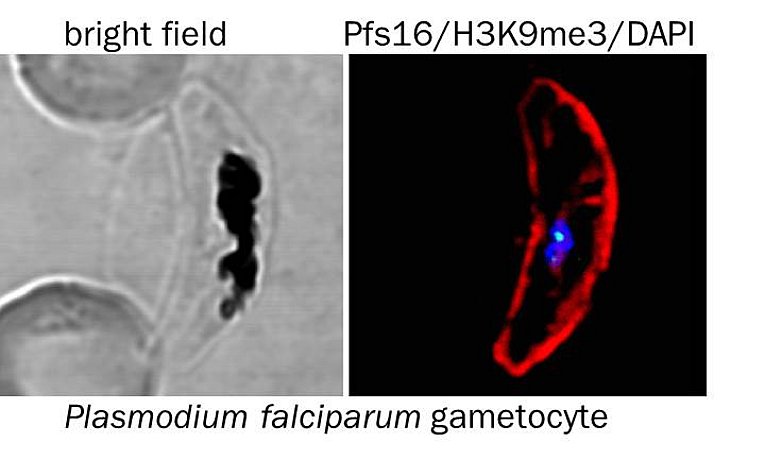
3) Sexual differentiation. Transmission of the malaria parasite to a new host requires the development of mature, sexual forms. The mechanisms that direct the lineage decision between sexual and asexual differentiation are poorly understood, but recent evidence indicates that epigenetic mechanisms are critical. Current drugs are inefficient in killing the sexual stages, resulting in prolonged transmission even after malaria disease is cured, moving the development of drugs that are effective against all blood stages into the focus of recent research efforts. We aim to create a detailed map of the epigenome and transcriptome of developing male and female gametocytes to deepen our understanding of the mechanisms that regulate commitment to and differentiation of both sexual forms.